Analytic Bijections¶
This tutorial demonstrates how to leverage the analytic bijections defined in bijx,
first in 1D and then in new radial flows in 2D.
from flax import nnx
import jax
import jax.numpy as jnp
import optax
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
import bijx
rngs = nnx.Rngs(0)
1D Example¶
We can learn onedimensional distributions by stacking sufficiently many scalar bijections. First, we’ll define a synthetic target distribution as an example.
@nnx.dataclass
class PolyTarget:
# Oscillatory Gaussian component coefficients
a1: nnx.Data[float] = 1.0 # Amplitude of sin term
w1: nnx.Data[float] = 5.0 # Frequency of sin
g1: nnx.Data[float] = 5.0 # Gaussian decay rate
# Cosine component
a2: nnx.Data[float] = 2.0 # Amplitude of cos term
w2: nnx.Data[float] = 10.0 # Frequency of cos
# Polynomial component
a4: nnx.Data[float] = -0.2
def __call__(self, x):
"""Compute unnormalized log density."""
# Handle both (batch,) and (batch, 1) shapes
x_flat = x.squeeze(-1) if x.shape[-1] == 1 else x
log_p = (
self.a1 * jnp.sin(self.w1 * x_flat) * jnp.exp(-self.g1 * x_flat**2)
+ self.a2 * jnp.cos(self.w2 * x_flat)
+ self.a4 * x_flat**4
)
return log_p
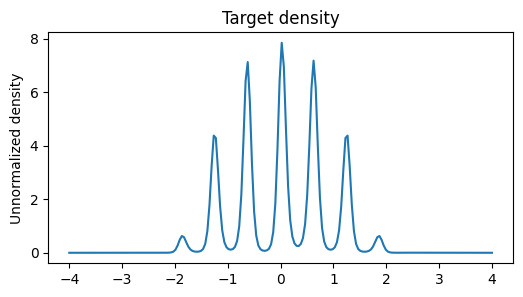
poly_target = PolyTarget()
plt.figure(figsize=(6, 3))
plt.title('Target density')
x = jnp.linspace(-4, 4, 200)
plt.plot(x, jnp.exp(poly_target(x)))
plt.ylabel('Unnormalized density')
plt.show()
Model¶
Instead of a for-loop over multiple bijections, we use bijx.ScanChain which chains the internal bijection using jax’s scan. The internal bijection is expected to have parameters with leading batch dimension, which is achieved by applying vmap over a number of random keys.
stack_size = 64
@nnx.split_rngs(splits=stack_size)
@nnx.vmap(in_axes=(0,), out_axes=0)
def create_bijection_stack(rngs):
# base = bijx.SinhConjugation(rngs=rngs)
# base = bijx.CubicRational(rngs=rngs)
base = bijx.CubicConjugation(rngs=rngs)
return bijx.ScanChain(base)
bijection = create_bijection_stack(nnx.Rngs(0))
model = bijx.Transformed(
bijx.IndependentNormal((), rngs=nnx.Rngs(0)),
bijection,
)
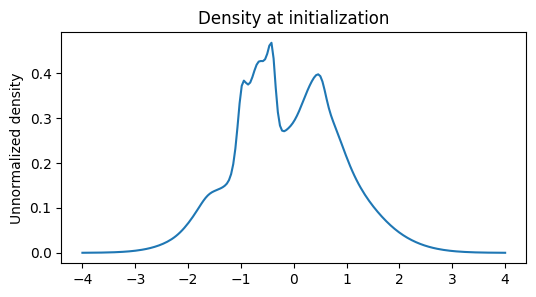
plt.figure(figsize=(6, 3))
plt.title('Density at initialization')
x = jnp.linspace(-4, 4, 200)
plt.plot(x, jnp.exp(model.log_density(x)))
plt.ylabel('Unnormalized density')
plt.show()
Training¶
# simple training setup
batch_size = 256
optimizer = nnx.Optimizer(
tx=optax.adam(1e-3),
model=model,
wrt=nnx.Param,
)
def loss_fn(model):
x, lq = model.sample((batch_size,))
lp = poly_target(x)
return -jnp.mean(lp - lq)
@nnx.jit
def update_step(model, optimizer):
loss, grads = nnx.value_and_grad(loss_fn)(model)
optimizer.update(grads=grads, model=model)
return loss
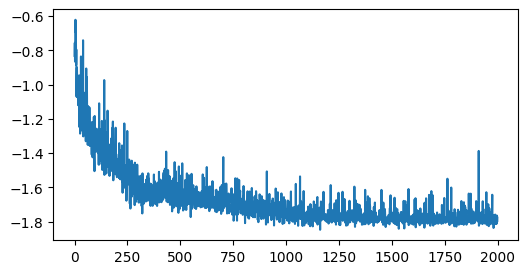
steps = 2000
losses = np.full(steps, np.nan)
for i in tqdm(range(steps)):
losses[i] = update_step(model, optimizer)
plt.figure(figsize=(6, 3))
plt.plot(losses)
plt.xlabel('Step')
plt.ylabel('Loss')
plt.show()
100%|██████████| 2000/2000 [00:05<00:00, 341.69it/s]
[<matplotlib.lines.Line2D at 0x1178bf0e0>]
Evaluation¶
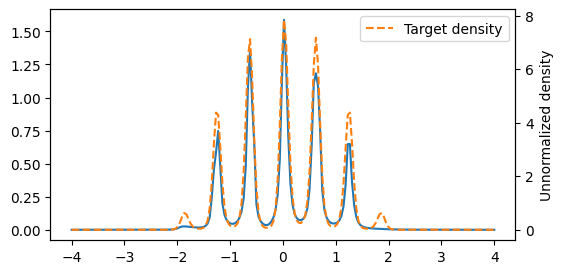
plt.figure(figsize=(6, 3))
x = jnp.linspace(-4, 4, 200)
plt.plot(x, jnp.exp(model.log_density(x)), label='Learned density')
plt.twinx()
plt.plot(x, jnp.exp(poly_target(x)), '--', label='Target density', color='C1')
plt.ylabel('Unnormalized density')
plt.legend()
plt.show()
Radial flows¶
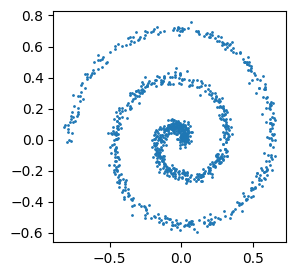
First we define a spiral distribution in 2D, this time by defining how to sample from it.
Target¶
@nnx.dataclass
class SpiralData:
n_turns: nnx.Data[float] = 2.5 # Number of spiral turns (5 * pi / (2 * pi) = 2.5)
radius_scale: nnx.Data[float] = 1.0 / 20.0 # Scaling factor for spiral radius
noise_scale: nnx.Data[float] = 0.02 # Gaussian noise added to spiral
is_normalized: bool = False # No density, so not normalized
@property
def event_shape(self) -> tuple[int, ...]:
return (2,)
def sample(self, batch_shape: tuple[int, ...] = (), *, rng):
"""Generate spiral samples using JAX-compatible operations."""
total_samples = int(np.prod(batch_shape))
# Generate parameter t uniformly from 0 to n_turns * 2 * pi
rng1, rng2 = jax.random.split(rng)
t = jax.random.uniform(rng1, (total_samples,), minval=0, maxval=self.n_turns * 2 * jnp.pi)
# Generate spiral coordinates
x = t * jnp.cos(t) * self.radius_scale
y = t * jnp.sin(t) * self.radius_scale
# Add Gaussian noise
noise = jax.random.normal(rng2, (total_samples, 2)) * self.noise_scale
# Combine and reshape
samples = jnp.stack([x, y], axis=1) + noise
return samples.reshape(*batch_shape, 2)
spiral_data = SpiralData()
samples = spiral_data.sample((1000,), rng=jax.random.key(0))
plt.figure(figsize=(3, 3))
plt.scatter(samples[:, 0], samples[:, 1], s=1)
plt.show()
Model architecture¶
First, we now want to transfrom radii, with respect to some trainable center. Thus, we must make sure that we map rays onto rays (i.e. preserve origin).
stack_size = 64
@nnx.split_rngs(splits=stack_size)
@nnx.vmap(in_axes=(0,), out_axes=0)
def create_bijection_stack(rngs):
# base = bijx.SinhConjugation(rngs=rngs)
# base = bijx.CubicRational(rngs=rngs)
base = bijx.CubicConjugation(rngs=rngs)
# now wrapped in ray transform
return bijx.RayTransform(bijx.ScanChain(base))
We will use a simple Fourier-series based coupling layer to make the radial transformation depend on the angle.
class FourierCondNet(nnx.Module):
"""Fourier-based conditioning network for angle-dependent parameters."""
def __init__(
self,
output_dim: int,
fourier_count: int = 3,
hidden_dim: int | None = None,
*,
rngs: nnx.Rngs,
):
assert fourier_count % 2 == 1, 'Fourier count must be odd'
self.kernel = bijx.nn.embeddings.KernelFourier(
fourier_count, val_range=(-jnp.pi, jnp.pi)
)
self.output_dim = output_dim
if hidden_dim is not None:
self.hidden = nnx.Linear(
fourier_count, hidden_dim,
use_bias=False, rngs=rngs,
kernel_init=nnx.initializers.normal(0.01),
)
final_in = hidden_dim
else:
final_in = fourier_count
self.superpos = nnx.Linear(
final_in, output_dim,
use_bias=False, rngs=rngs,
kernel_init=nnx.initializers.normal(0.01),
)
def __call__(self, x_hat):
"""Map unit vectors to bijection parameters.
Args:
x_hat: Unit vectors of shape (..., 2).
Returns:
Parameters of shape (..., output_dim).
"""
theta = jnp.atan2(x_hat[..., 1], x_hat[..., 0])
features = self.kernel(theta)
# Regularize feature magnitudes: suppress high frequencies
freq_count = (self.kernel.feature_count - 1) // 2
freq_indices = jnp.concatenate([
jnp.arange(1, freq_count + 1), # for sin terms
jnp.arange(1, freq_count + 1), # for cos terms
jnp.array([1.0]) # for constant term
])
features = features / freq_indices
if hasattr(self, 'hidden'):
features = self.hidden(features)
return self.superpos(features)
def create_radial_layer(rngs):
base = create_bijection_stack(rngs)
cond_net = FourierCondNet(
output_dim=bijx.ModuleReconstructor(base).params_total_size,
fourier_count=7,
rngs=rngs
)
layer = bijx.RadialConditional(
scalar_bijection=base,
cond_net=cond_net,
center=jnp.array([-0.5, -1.]),
scale=jnp.zeros(2),
rngs=rngs
)
return layer
model = bijx.Transformed(
bijx.IndependentNormal((2,), rngs=rngs),
create_radial_layer(rngs)
)
Training¶
# simple training setup
batch_size = 512
optimizer = nnx.Optimizer(
tx=optax.adam(1e-2),
model=model,
wrt=nnx.Param,
)
def loss_fn(model):
x = spiral_data.sample((batch_size,), rng=model.rngs())
lq = model.log_density(x)
return -jnp.mean(lq)
@nnx.jit
def update_step(model, optimizer):
loss, grads = nnx.value_and_grad(loss_fn)(model)
optimizer.update(grads=grads, model=model)
return loss
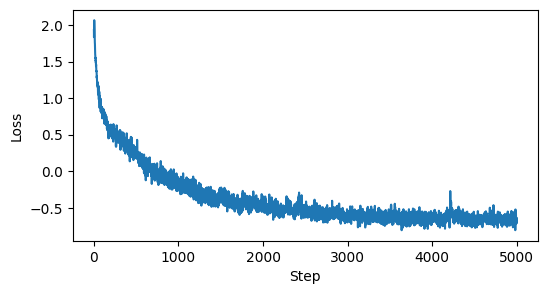
steps = 5000
losses = np.full(steps, np.nan)
for i in tqdm(range(steps)):
losses[i] = update_step(model, optimizer)
plt.figure(figsize=(6, 3))
plt.plot(losses)
plt.xlabel('Step')
plt.ylabel('Loss')
plt.show()
100%|██████████| 5000/5000 [00:33<00:00, 148.29it/s]
Evaluation¶
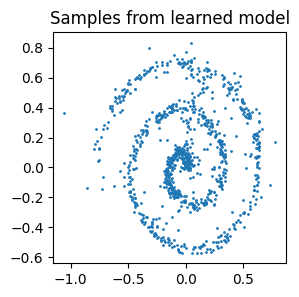
x, _ = model.sample((1000,))
plt.figure(figsize=(3, 3))
plt.title('Samples from learned model')
plt.scatter(x[:, 0], x[:, 1], s=1)
plt.show()